John Appleyard, updated 21st April 2020
Disclaimer: I trained as a physicist to PhD level many years ago, and my career required good numeracy. However, I have no special expertise in epidemiology or statistics.
This version is no longer current – please see
https://fortran.uk/using-published-covid-19-data-to-deduce-real-infection-rates/
Introduction
In this note, I will show that a simple model of linking testing with confirmed cases and deaths is confirmed by analysis of published data. The resulting equation can be extrapolated to predict true infection rates. It is argued that, although the model must break down for high infection rates, the way in which it breaks down can be anticipated and incorporated into a larger framework.
The Model for Covid-19 Testing
If testing were random, we would expect the number of cases found (c) to be proportional to the number of tests (t), and the proportion of the population that is infected. The second factor is unknown, but the number of deaths (d) is probably a reasonable proxy, so we have:
c ∝ d.t
A better proxy for the infection rate now might be the death rate in a few days time, but initially, we will use the simpler model.
In the early stages of the pandemic, testing is not random, but is designed to find as many cases as possible. It focuses on a small part of the population comprising the contacts of existing cases, medical staff, and those with symptoms. If the size of that reduced population is proportional to c, then the number of cases found will be
c ∝ d.t/c
c ∝ sqrt(d.t)
In effect, testing is concentrated on a bubble of contacts surrounding each case, and for small c, these bubbles are isolated. As c increases, the bubbles increasing overlap, and eventually encompass the entire population. At that point testing effectively becomes random, with c ∝ d.t.
To summarize, the hypothesis is that c should vary with sqrt(d.t) for small c, and with d.t for large c.
Comparison with Published Data
The data, downloaded from worldometers.info on the 14th April 2020, comprises 3 variables for each of 110 countries with more than 100 cases, and at least one death. China is excluded because testing data is not available. The variables are the number of confirmed cases (c), deaths (d), and tests (t) per million of population.
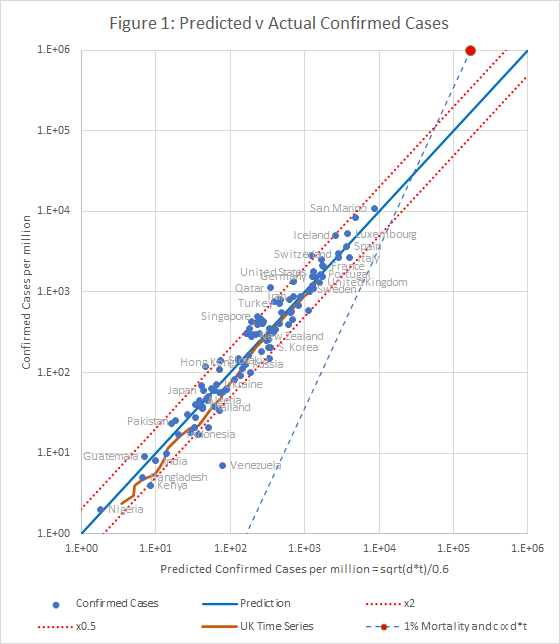
Figure 1 shows that the data fits very well to the equation
c = 1.67 * sqrt(d*t)
The prediction spans almost 4 orders of magnitude, but even so, almost all countries are within the tramlines, which indicate a factor of 2 either side of the predicted value.
There are many reasons to expect data from different countries to differ, including:
- Different health systems – some primitive, and others advanced
- Overloading makes even advanced health systems less effective
- Different demographics – Covid-19 is worse for older populations
- Time lags – e.g. confirmed cases may not survive, and reports may be delayed
- Different criteria for counting deaths
- Political massaging of published data
The conclusion is that all of these factors combine to a factor of up to about 2.
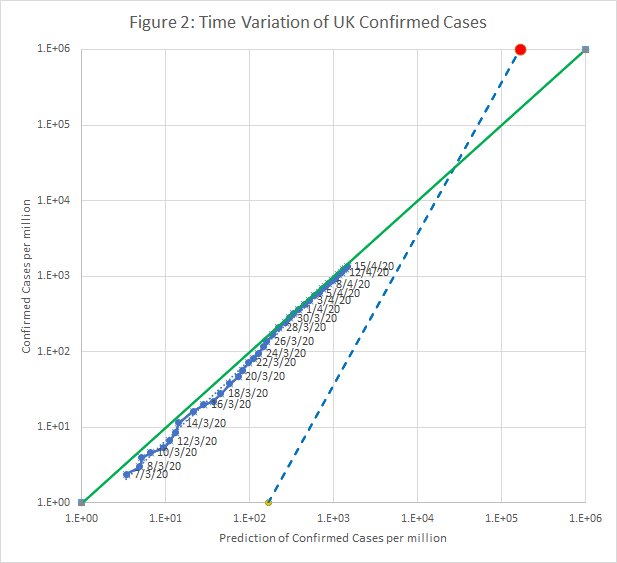
UK Time Series
Historic data for the UK was downloaded from https://github.com/tomwhite/covid-19-uk-data/blob/master/data/covid-19-indicators-uk.csv
Figure 2 shows how the model fits this data. This time-line is also superimposed, as an orange trace, on Figure 1.
This line is also superimposed, as an orange trace, on Figure 1.
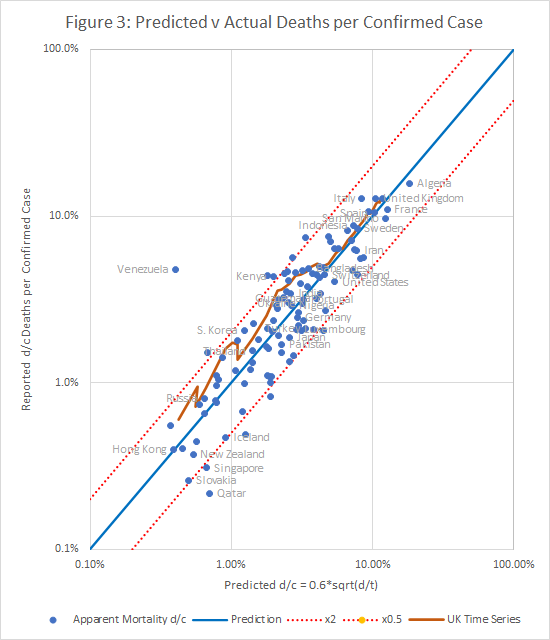
Matching Case Fatality Rates (d/c)
Case fatality rates are an obvious but misleading measure, which is often used wrongly, when comparing different countries. In general terms, it is known that countries that test extensively tend to have lower case fatality rates, but the model described here allows us to quantify that relationship.
Figure 3 shows that a good fit to the observed apparent fatality rate, d/c, can be obtained using the equation:
d/c=0.6*sqrt(d/t)
Once again, only a few countries diverge from the prediction by more than a factor of 2 (indicated by the tramlines). It is possible to obtain slightly better fit metrics by multiple regression, but the difference is small, and not obvious on a scatter plot.
Predictions from the Model, and How to Extend it
If we assume c = t = 1 million (i.e. the entire population is infected), the model predicts mortality as 36%. This is clearly wrong, and suggests that as the proportion infected increases, the curve in Figures 1 and 2 turns up so that the mortality at saturation is about 1%. This point is indicated by a red spot.
The dashed line projected down from the red spot represents the cases that would be found with random testing if the true infection fatality rate were 1%.
As the number of cases and tests increases to include a significant proportion of the population, it’s likely that testing has to be more random. If the true mortality rate is 1%, that suggests that the actual course of the pandemic veers away from the straight line extrapolation of the model and on to the dashed line representing random testing.
The point at which the two lines cross is determined by
c = 1.67*sqrt(d*t) = (d*t)/(f*1E6)
c = 2.79E6*f
where f is the true infection mortality rate. For f = 1%, that is when the reported cases, c are 2.79% of the population.
This is a gentle deviation that occurs in a region for which we have little data.
The model can also be used to predict the true number of infections per million (c0) at any given time. c varies with the square root of the number of tests, so we can get an estimate of c0 using
c0 = c . sqrt(1E6 / t)
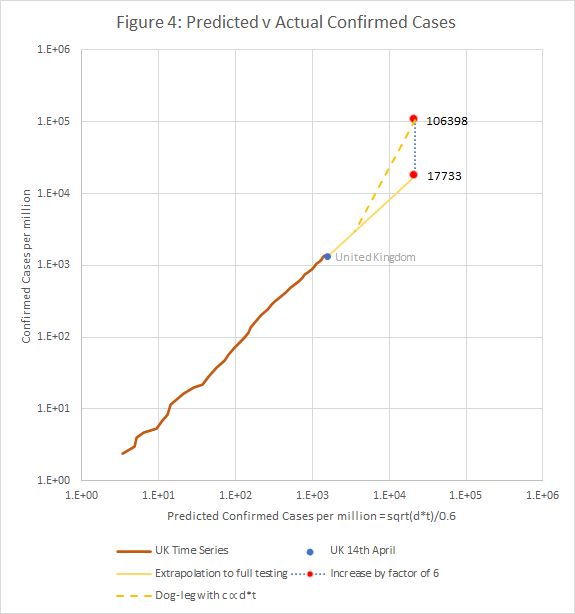
For the UK, on 14th April, that is about 1.8% of the population. However, that is probably an underestimate, as we suspect that the model may break down as the number of tests is increased towards saturation. In the case where c = t = 1E6, a factor of 6 increase in the extrapolated value of c was required to bring the predicted mortality down from 36% to 1%. In general the factor is 0.6/sqrt(f).
If the same were true for lower infection rates, we’d have to increase the computed value of c0 by a factor of 6, meaning that around 10% of the population has been infected. However it is not clear whether, the model breaks primarily because of the high infection rate, or the high testing rate, or a combination. The true figure is most likely between 1 and 0.6/sqrt(f).
Figure 4 shows how the transition from the model to a regime with c ∝ d.t might work.
You can download an Excel spreadsheet containing the data, workings and charts here.